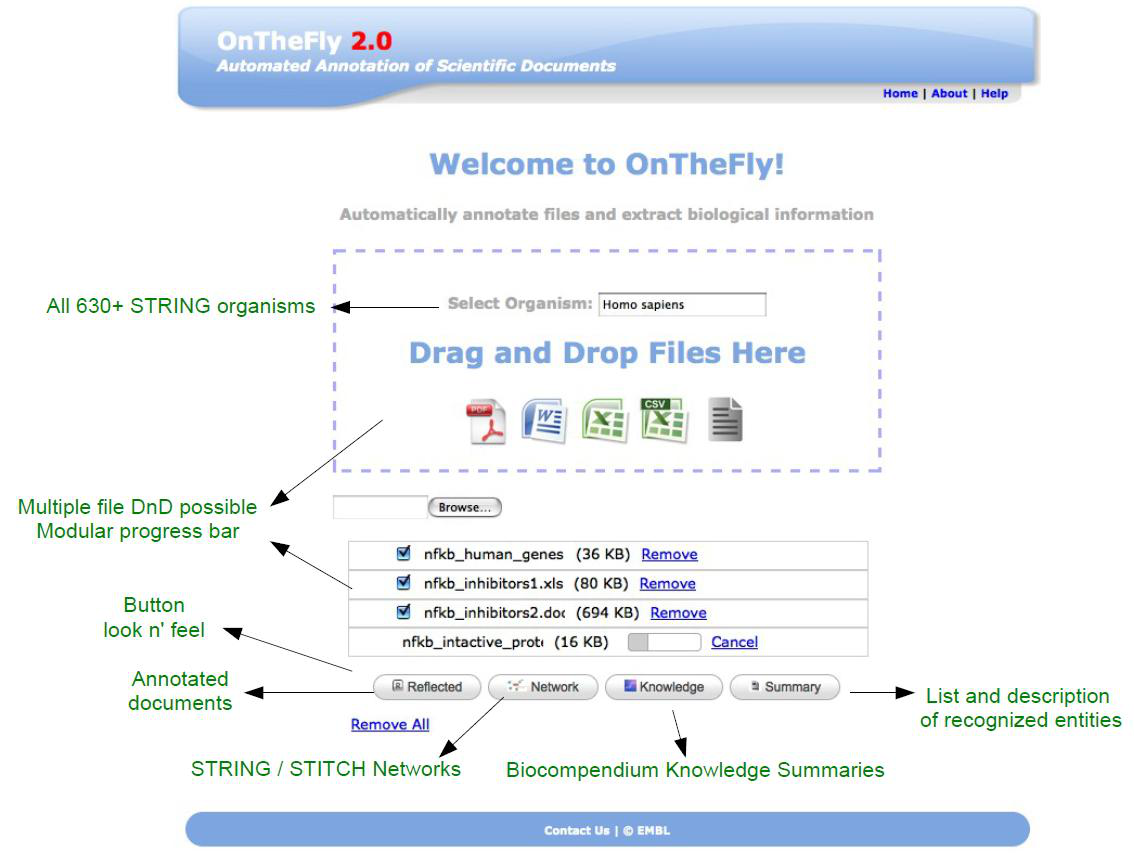
OnTheFly 2.0 interface: After selecting their organism of interest (from an autocomplete list), researchers may just Drag and Drop files into OnTheFly. The documents will be automatically annotated by Reflect and further analysis will be possible.
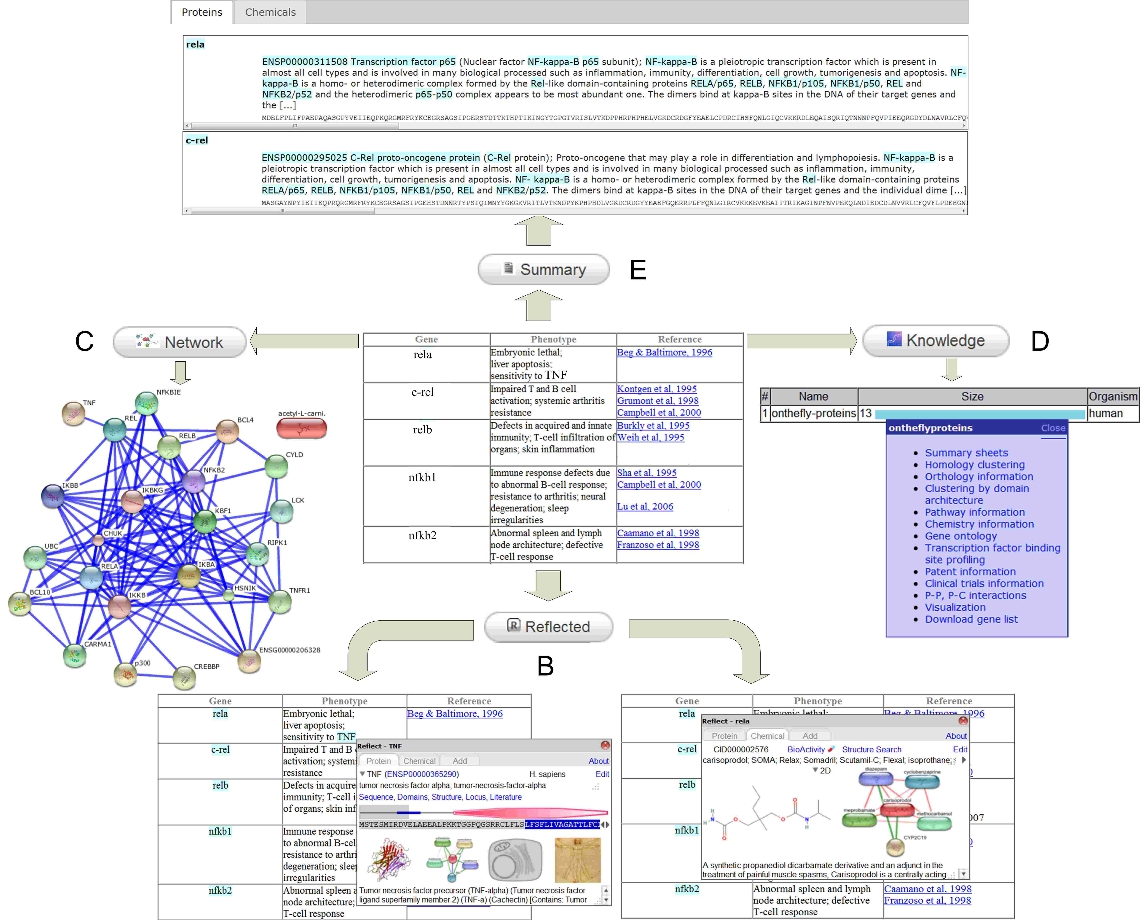
OnTheFly 2.0 main functionality: A) A document file containing an Nf-kB regulatory network description B) Reflected Button Document annotation through the Reflect service C) Network Button: Protein(s)/chemical(s) interaction network generation through the STITCH database D) Knowledge Button: Collection of relevant knowledge from public resources via the Biocompendium service E) Summary Button: Enriched summaries containing short descriptions about the bioentities of a set of documents
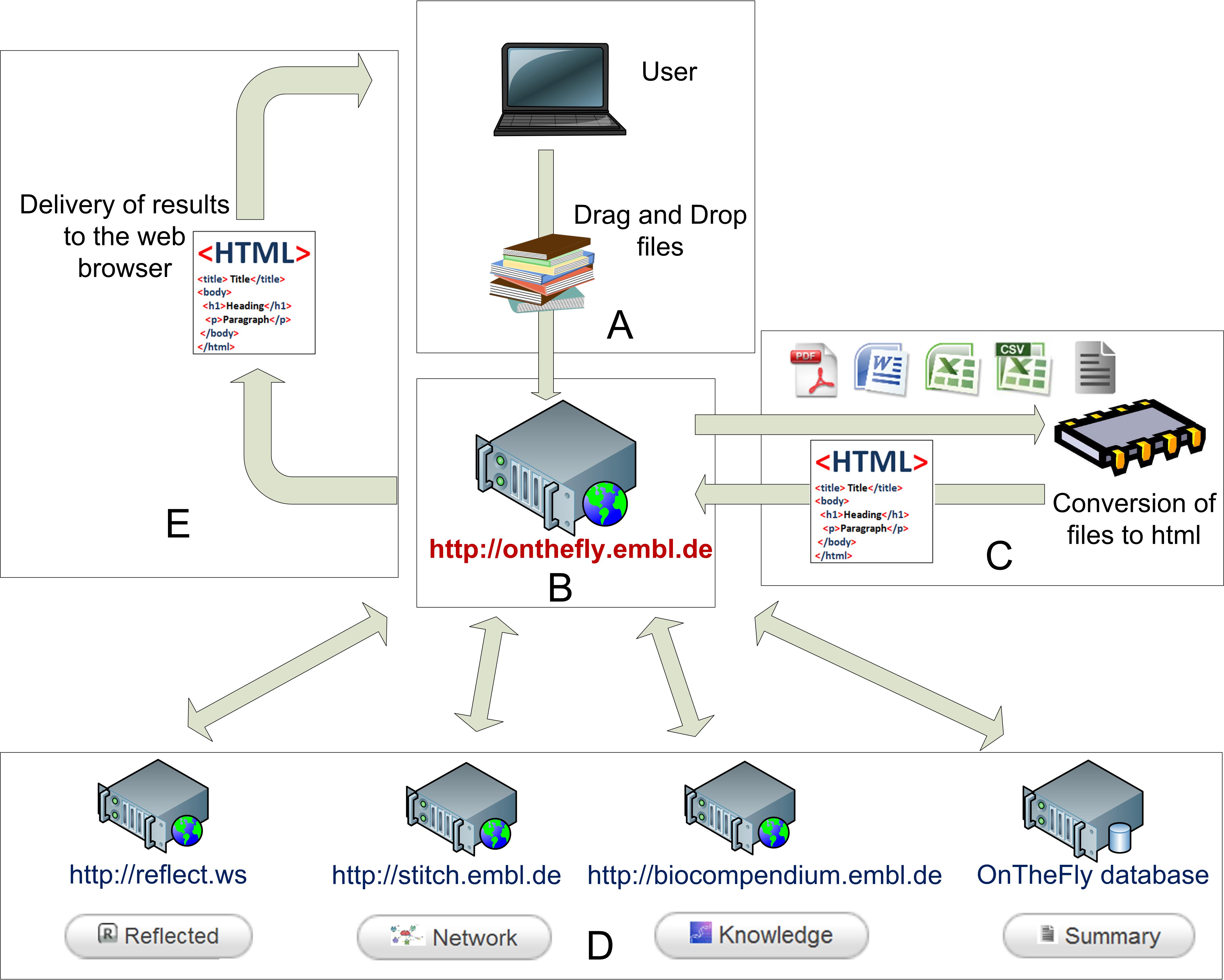
OnTheFly 2.0 architecture: An overview of the architecture of OnTheFly. OnTheFly central sever in Greece and its information exchange with other servers (Reflect server based in Dennmark, Stitch server in Germany and Biocompendium server in Luxembourg)
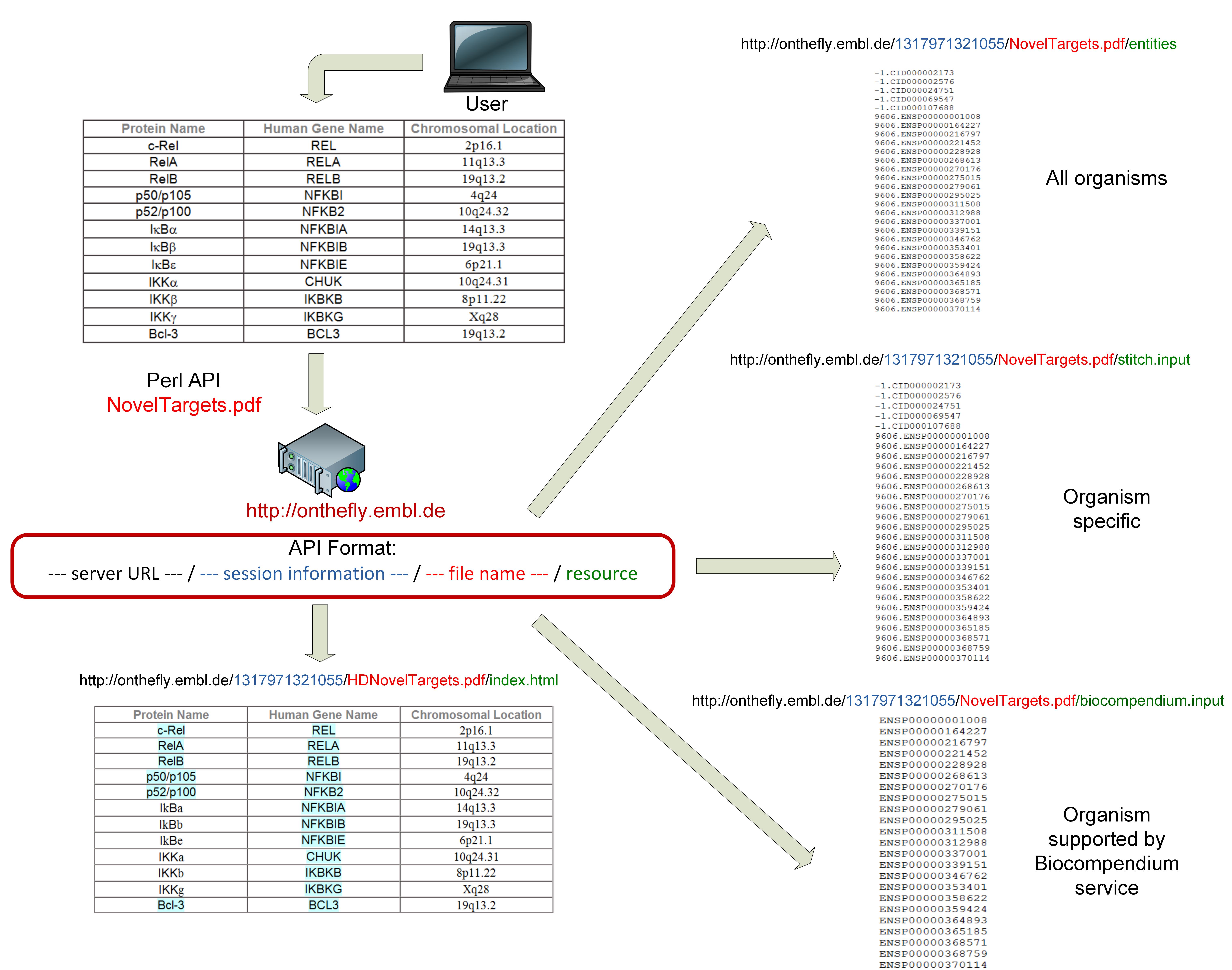
- "http://onthefly.embl.de/sessionID/filename/entities"
- "http://onthefly.embl.de/sessionID/filename/stitch.input"
- "http://onthefly.embl.de/sessionID/filename/biocompendium.input".
- "http://onthefly.embl.de/Session"
- "http://onthefly.embl.de/Organisms"
A Perl API can be downloaded here
The list of organisms with their taxonomy Ids is available here